gbspy.fits module
Provide commonly used fitting profiles.
- class gbspy.fits.FitConfig(fun, **kwargs)
Bases:
objectFit configuration object
Holds arguments to call
scipy.optimize.curve_fit()with.- fun
Target profile to fit as a callable object.
- Type
callable
- update_fit_opts(**new_opts)
Update kwargs options
This will call the underlying
dict.update()on the options stored by the class. Existing options will be replaced, additional arguments will create new options.- Parameters
**new_opts (dict, optional) – Options to add or update, which must be accepted by
curve_fit().- Returns
The updated FitConfig object.
- Return type
- gbspy.fits.ffun_double_exp(x, q0, x0, L_left, L_right)
Double exponential profile
This profile is characterized as
\[ \begin{align}\begin{aligned}f(x) = q_0 \exp(-(x-x_0)/L(x)),\\\begin{split}L(x) = \begin{cases} L_{\text{left}}, & \text{if}\ x<x_0 \\ L_{\text{right}}, & \text{otherwise} \end{cases}\end{split}\end{aligned}\end{align} \]- Parameters
- Returns
The profile evaluated at
x.- Return type
array_like
- gbspy.fits.ffun_eich2011(x, q0, L, S, qb)
Convolution of an exponential profile and a Gaussian function
This profile is proposed by Eich et al. as the result of a competition between parallel plasma transport along a divertor leg and perpendicular diffusion (leakage) into the private flux region, assuming an exponential profile at the divertor entrance. A convolution between an exponential profile and exponential is proposed to model this process:
\[f(x) = \frac{q_0}{2} \exp\left(\frac{S^2}{4L^2} - \frac{x}{L}\right) \mathrm{erfc}\left(\frac{S}{2L} - \frac{x}{S}\right) + q_{\text{BG}}\]where \(S\) the width of the Gaussian and \(L\) the decay length of the exponential profile. See equation (2) in Eich et al. (2011).
- Parameters
- Returns
The profile evaluated at
x.- Return type
array_like
- gbspy.fits.ffun_exp(x, q0, L)
Exponential profile
Defined by
\[f(x) = q_0 \exp(-x/L)\]
- gbspy.fits.ffun_gaussian(x, q0, x0, L)
Gaussian function
Defined as:
\[f(x) = q_0 \exp\left(\frac{-(x-x_0)^2}{2L^2}\right)\]
- gbspy.fits.ffun_linear(x, a, b)
Linear profile
Returns the result of evaluating
\[f(x) = ax + b\]
- gbspy.fits.fit(f, xdata, ydata, return_func=None, **kwargs_scipy)
Non-linear least square fitting wrapper
This function wraps calls to
scipy.optimize.curve_fit(). In addition to scipy’s routine, this function accepts a string as first argument to reference commonly used fitting profiles.When the first argument is a callable, this routine behaves like like scipy’s
curve_fit().When the first argument is a string:
optional arguments accepted by scipy’s routine (such as
boundsorp0) may be set automatically. You can override their values by passing them directly tofit()yourself. You can see which parameters are set by callingget_fit_configuration(),unless specified otherwise, the fitting function is also returned (as the last return value).
- Parameters
f (callable or str) –
If string, the following options are accepted:
"linear":Linear function,"exp":Exponential decay,"doubleexp":Double exponential decay,"gaussian":Gaussian function,"eich2011": seeffun_eich2011().
return_func (bool, optional) – Whether to return the fitting function.
Trueiffis a string,Falseotherwise.kwargs_scipy (dict, optional) – Any keyword argument accepted by
curve_fit().
- Returns
popt (array) – See
curve_fit().pcov (2-D array) – See
curve_fit().func (callable) – The function that was used to fit the curve. This argument is only returned when the first argument is a string, or when explicitely enabled by the caller.
Examples
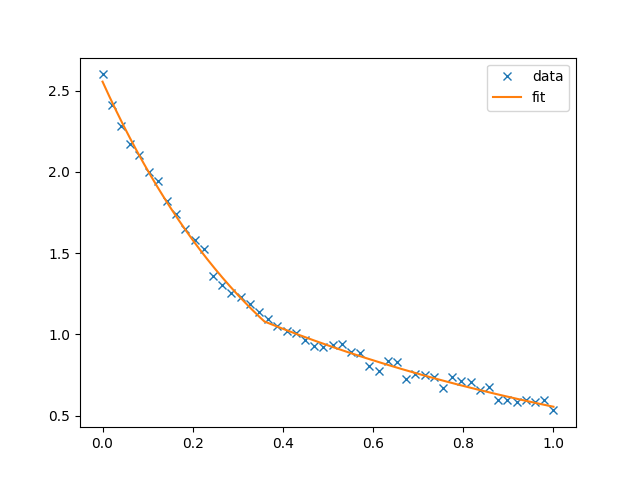
Generate a noisy double exponential profile: two decay lengths of
0.4and0.95are considered. The transition between the decay lengths occurs atx=0.3678:In [1]: import gbspy.fits as fits In [2]: x = np.linspace(0, 1, 50) In [3]: y_true = fits.ffun_double_exp(x, 1.0, 0.3678, 0.4, 0.95) In [4]: noise = 1e-1 * np.random.random((50, )) In [5]: y = y_true + noise
Fit the curve. Here, we provide a reasonable guess of
0.5at which the decay length changes,In [6]: params, cov, ffun = fits.fit("doubleexp", x, y, p0=[1.0, 0.5, 1.0, 1.0]) In [7]: print( ...: ( ...: f"First decay length: {params[2]!r}\n" ...: f"Second decay length: {params[3]!r}\n" ...: f"Transition at: {params[1]!r}" ...: ) ...: ) ...: First decay length: 0.4161962043210802 Second decay length: 0.9621985264070205 Transition at: 0.35874387625916493
This yields a reasonable estimate of the parameters. We can also plot the result of the fit with
In [8]: import matplotlib.pyplot as plt In [9]: plt.figure() Out[9]: <Figure size 640x480 with 0 Axes> In [10]: plt.plot(x, y, "x", label="data") Out[10]: [<matplotlib.lines.Line2D at 0x7f0ac485a940>] In [11]: xhres = np.linspace(0, 1, 2000) In [12]: plt.plot(xhres, ffun(xhres, *params), label="fit") Out[12]: [<matplotlib.lines.Line2D at 0x7f0ac485abb0>] In [13]: plt.legend() Out[13]: <matplotlib.legend.Legend at 0x7f0ac48c4cd0>
- gbspy.fits.get_fit_configuration(name, x, y)
Returns fitting options based on input data
Given the name associated with a given profile, returns both a callable for this profile and fitting options. The fitting options must be accepted by
scipy.optimize.curve_fit().- Parameters
name (str) – Name associated to the profile that must be fit (for example,
"linear").x (array_like or object) – Data points to be fitted, accepted by scipy’s
curve_fit().y (array_like or object) – Data points to be fitted, accepted by scipy’s
curve_fit().
- Returns
An object storing the callable associated to the requested profile and options to pass to
curve_fit().- Return type