|
GBS
|
 |
GBS
|
This page explains how to obtain and compile the GBS code
The GBS code is version controlled using git and the central repository is hosted on the c4science.ch platform. To access it, you must create a c4science account and then ask for access to the source code (precising your c4science username) by email to paolo.ricci@epfl.ch.
Once you have been granted access, you can clone the GBS repository using:
or
Depending on the chosen protocol (https or ssh) you will need to setup either a VCS password or a public ssh key.
The GBS code relies on many different libraries. The following are mandatory:
For the solvers, you can choose one of the following:
One other in-house library, futils, is packaged with GBS and compiled automatically.
On Marconi, there are a few extra steps to perform before being able to compile GBS using CMake. First of all, here are the modules you need to load:
Note that it is important to load the PETSc module without the _complex in the name. Otherwise, it will also provide an FFTW interface, which will cause problems.
Then, add the following in your .bashrc file on Marconi:
The first line is used to tell CMake where to find PETSc (it is a missing configuration on their module) and the second line tells CMake to use Metis and MUMPS provided by the PETSc module. For some reason, it does not work with the standalone modules provided on Marconi.
This concludes the configuration specific to Marconi. You can now proceed as explained at the beginning of Setting up CMake.
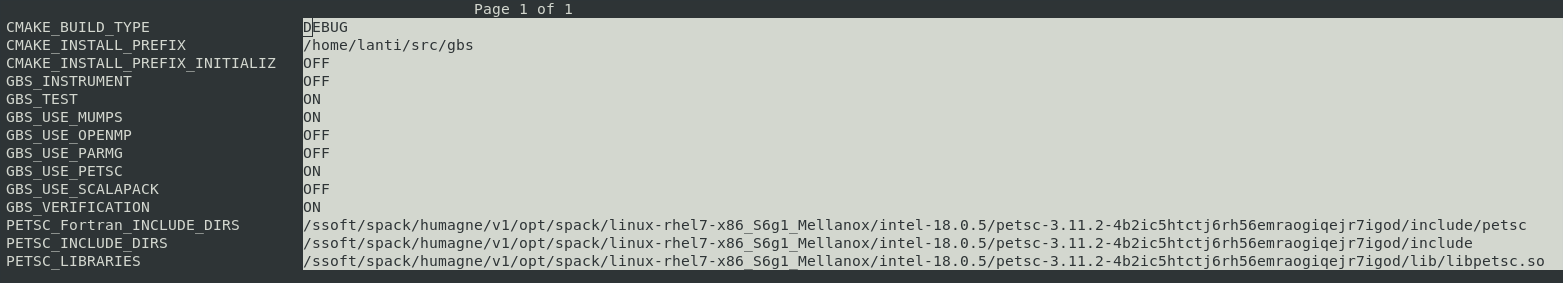
The configuration and compilation of the code is done using CMake and Make. In the GBS root directory, the following commands are used to create a directory and setting up CMake:
CMake will automatically search for the required libraries and set up a makefile. Once the configuration is done, you can either compile the code as is using
or customize the compilation options using
This will open a GUI with a summary of the different options available for GBS